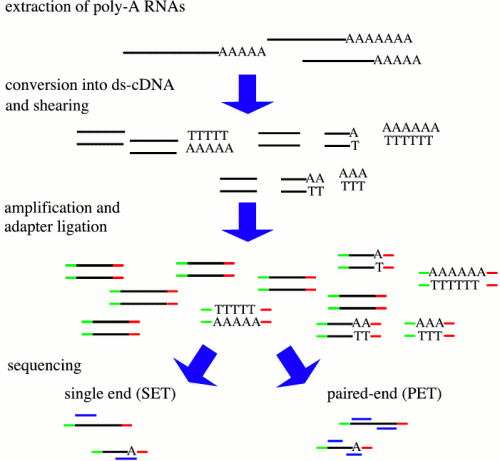
RNA-Seq is a new protocol for the analysis of the RNA content of a cell. Here, we are primarily interested in the polyadenylated RNA content, mostly mRNAs, but there are variatons for extracting other small RNAs like microRNAs.
As a first step in the protocol only polyadenylated RNAs are extracted. The exact protocol varies mostly in the aspects of shearing the cDNA (e.g. nebulization or sonication), the strategy of amplification (before/after fragmentation ...), and the type of sequencing (i.e. single end or paired end sequencing). See below for references of publications where such data sets have been analyzed with different focuses and different sequencing platforms have been used for RNA-sequencing (1. Illumina Genome analyzer, 2. Applied Biosystems Solid, 3. Roche 454 Life Sciences).
References
-
Sultan,M.*, Schulz,M.H.*, Richard,H.*, Magen,A., Klingenhoff,A., Scherf,M., Seifert,M., Borodina,T., Soldatov,A., Parkhomchuk,D., Schmidt,D., O'Keeffe,S., Haas,S., Vingron,M., Lehrach,H., Yaspo,M.L. (2008)
A Global View of Gene Activity and Alternative Splicing by Deep Sequencing of the Human Transcriptome
Science, 321(5891):956-960
-
Nicole Cloonan*, Alistair R R Forrest*, Gabriel Kolle*, Brooke B A Gardiner, Geoffrey J Faulkner, Mellissa K Brown, Darrin F Taylor, Anita L Steptoe, Shivangi Wani, Graeme Bethel, Alan J Robertson, Andrew C Perkins, Stephen J Bruce, Clarence C Lee, Swati S Ranade, Heather E Peckham, Jonathan M Manning, Kevin J McKernan & Sean M Grimmond
Stem cell transcriptome profiling via massive-scale mRNA sequencing
Nature Methods, July 2008, Volume 5, No 7, pages 613 - 619
-
J. CRISTOBAL VERA*, CHRISTOPHER W. WHEAT*, HOWARD W. FESCEMYER, MIKKO J. FRILANDER, DOUGLAS L. CRAWFORD, ILKKA HANSKI and JAMES H. MARDEN
Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing
Molecular Ecology, 2008, Volume 17 Issue 7, pages 1636 - 1647
*joint first authors

